Difference between revisions of "The Biolibrarian Proposal"
| Line 52: | Line 52: | ||
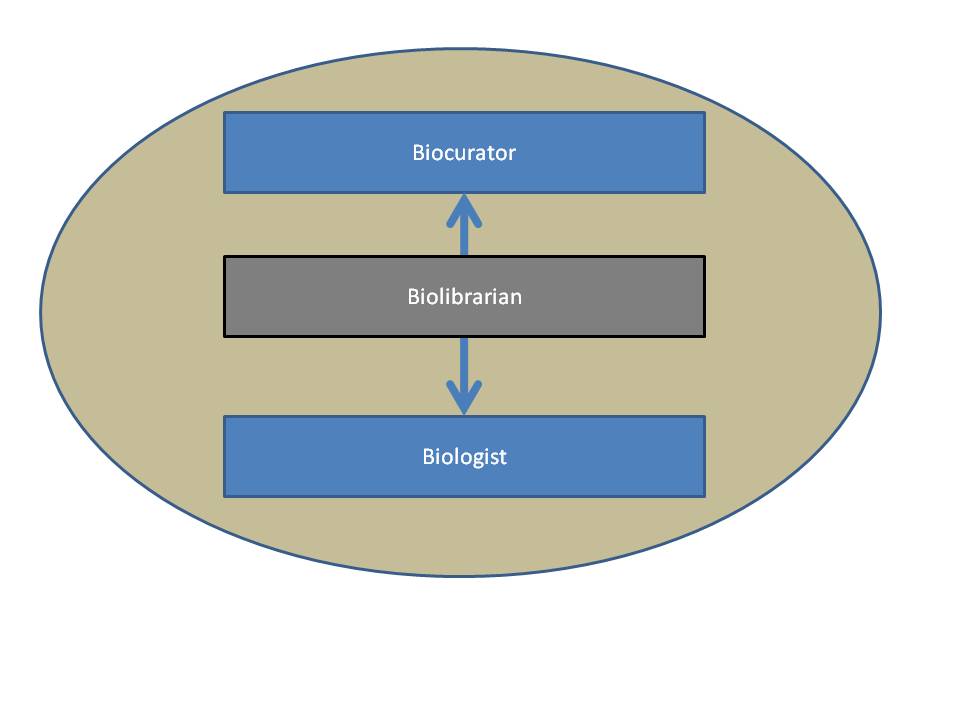
Biologists, BioLibrarians and Biocurators are all Bioinformaticians. Each group both consumes and produces biological information. Biologists are the primary producers of biological information. Biocurators care for some specific subset of biological information associated with a specific database. Biolibrarians care for the transfer of knowledge between biologists and databases (Biocurators) regardless of the focus of the biologist or the database. Bioinformatics encompasses all aspects of this process of data production and consumption. The defining and differentiating aspects of the Biolibrarian are: | Biologists, BioLibrarians and Biocurators are all Bioinformaticians. Each group both consumes and produces biological information. Biologists are the primary producers of biological information. Biocurators care for some specific subset of biological information associated with a specific database. Biolibrarians care for the transfer of knowledge between biologists and databases (Biocurators) regardless of the focus of the biologist or the database. Bioinformatics encompasses all aspects of this process of data production and consumption. The defining and differentiating aspects of the Biolibrarian are: | ||
| − | 1) BioLibrarians are non-partisan with respect to databases (as opposed to biocurators): this means that biolibrarians will choose databases to submit data to based on their their appropriateness to the data at hand to be archived. These | + | 1) BioLibrarians are non-partisan with respect to databases (as opposed to biocurators): this means that biolibrarians will choose databases to submit data to based on their their appropriateness to the data at hand to be archived. These databases may include protein databases such as UniProt, EntrezGene or the Gene Ontology for functional annotation of gene products or the [http://ocelot.uio.no/nesys/ FACCS database] for cataloging of the structure and structure-function relationships in the cerebro-cerebellar system. Biolibrarians may review and recommend databases and be involved in discussions around the use of and changes to database representations and controlled vocabularies. |
| − | 2) Biolibrarians do not write code (as opposed to bioinformaticians). | + | 2) Biolibrarians do not write code and are non-partisan with respect to development groups (as opposed to bioinformaticians). This means that Biolibrarians are primarily users of databases and associated software. They may review existing software or even tender the creation of new software either locally or internationally . Here, their knowledge of domain representation and query use-cases in a specific area would be used to shape requirements. However, Biolibrarians would not use or endorse software solely because it is produced and maintained by a research group at their local university. |
| − | 3) Biolibrarians facilitate submission of data from biologists to databases. | + | 3) Biolibrarians facilitate submission of data from biologists to databases. This is an active (not a passive) mandate. Biolibrarians would actively seek out and engage local researchers to collect and curate data that is either specific to the local research group or that is the specialization of the local Biolibrarian group. |
Biolibrarians may already exist as part of bioinformatics service groups if they facilitate both data use ''and'' submission without regard to a specific data type or database. | Biolibrarians may already exist as part of bioinformatics service groups if they facilitate both data use ''and'' submission without regard to a specific data type or database. | ||
Revision as of 23:55, 2 April 2009
Contents
- 1 The survey
- 2 The Biolibrarian
- 3 What is the Biolibrarian proposal?
- 4 The Biolibrarian's role defined
- 5 How do Biolibrarians differ from Biocurators and Bioinformaticians?
- 6 Guiding principals of the Biolibrarian proposal
- 7 Existing problems addressed by the Biolibrarian proposal
- 8 The proposal in brief
- 9 How to add your university to this survey
- 10 Related sites
The survey
Please take the Biolibrarian proposal survey by visiting http://www.surveymonkey.com/s.aspx?sm=LubRkhzbX9a7e8aPGKGevQ_3d_3d
It's only six questions and can be filled out in less than five minutes.
The Biolibrarian
The Biolibrarian takes ownership for the transfer of information between biologists and biological databases.
What is the Biolibrarian proposal?
The Biolibrarian is a proposed new infrastructure position at university libraries around the world.
A Biolibrarian is trained in the use of biological databases and acts to
1) help biologists locate and use biological databases
2) help biologists submit data and feedback to biological databases
It is envisioned that molecular biologists could meet with a Biolibrarian in the same way that they meet with and use the services of a librarian. For example, the Biolibrarian could help a molecular biologist researcher to locate pathways, complexes and interactions that their molecules of interest are involved in.
In addition, the Biolibrarian could help biologists users to locate and submit data and feedback to biological databases.
For example, the Biolibrarian would be trained in the use of state of the art text mining tools to help researchers locate data for their molecules of interest in abstracts and full-text research articles. They could then help researchers enter verified information from full-text articles into curated databases where it would be available to researchers around the world that were querying for information on these same molecules.
In this role, the Biolibrarian would act as a broker between local researchers and database curators. This would apply to those databases that are set up to accept feedback and entries from external sources. Biolibrarians would also become local brokers that provide feedback on databases, interfaces and associated search tools.
Would you support such a service at your local university library? Do you have comments on this proposal? Follow the link above and take our survey.
You can read a synopsis of the proposal below.
Comments and suggestions are welcome. We are also interested in learning about similar proposals or projects that are already in place. Please email ian.donaldson@biotek.uio.no. If you would like to add your name to this wiki page in support of this application, please send a brief email.
The Biolibrarian's role defined
Biolibrarians:
act as brokers between biologists and databases: this means that they facilitate interaction between local biologists and biological databases. This is a two-way process that includes helping bioloigists find and use biological databases AND helping biologists submit data and feedback to biological databases.
facilitate retrieval of biological information: biolibrarians would be trained with specialized knowledge of major biological databases, that includes their data representations, data exchange mechansisms with other databases, and the controlled vocabularies and ontologies that they employ. These are all important aspects of being able to find, use and interpret biological data. Examples of major biological databases include databases like GenBank, UniProt, the Saccharomyces Genome Database, Genetic Association Database, Gene Ontology Database, The Protein Databank, The Eukaryotic Linear Motif Database, GAPScreener, IntAct, BioModels Database and Reactome. These are just a few of the hundreds of biological databases that may be of relevance to local biologists MetaDB.
facilitate submission of data to biological databases: Each of the databases listed above will accept record submissions and/or feedback from biologist users. The quality of these databases is dependent on this relationship and many of these databases face critical shortages in curation personnel. The Biolibrarian's role is to to actively seek out and engage biologists in submitting data and feedback to these databases. The sheer number of databases, data models, and controlled vocabularies employed by these databases requires a specialist intermediary to facilitate this exchange of information.
How do Biolibrarians differ from Biocurators and Bioinformaticians?
Biologists, BioLibrarians and Biocurators are all Bioinformaticians. Each group both consumes and produces biological information. Biologists are the primary producers of biological information. Biocurators care for some specific subset of biological information associated with a specific database. Biolibrarians care for the transfer of knowledge between biologists and databases (Biocurators) regardless of the focus of the biologist or the database. Bioinformatics encompasses all aspects of this process of data production and consumption. The defining and differentiating aspects of the Biolibrarian are:
1) BioLibrarians are non-partisan with respect to databases (as opposed to biocurators): this means that biolibrarians will choose databases to submit data to based on their their appropriateness to the data at hand to be archived. These databases may include protein databases such as UniProt, EntrezGene or the Gene Ontology for functional annotation of gene products or the FACCS database for cataloging of the structure and structure-function relationships in the cerebro-cerebellar system. Biolibrarians may review and recommend databases and be involved in discussions around the use of and changes to database representations and controlled vocabularies.
2) Biolibrarians do not write code and are non-partisan with respect to development groups (as opposed to bioinformaticians). This means that Biolibrarians are primarily users of databases and associated software. They may review existing software or even tender the creation of new software either locally or internationally . Here, their knowledge of domain representation and query use-cases in a specific area would be used to shape requirements. However, Biolibrarians would not use or endorse software solely because it is produced and maintained by a research group at their local university.
3) Biolibrarians facilitate submission of data from biologists to databases. This is an active (not a passive) mandate. Biolibrarians would actively seek out and engage local researchers to collect and curate data that is either specific to the local research group or that is the specialization of the local Biolibrarian group.
Biolibrarians may already exist as part of bioinformatics service groups if they facilitate both data use and submission without regard to a specific data type or database.
Guiding principals of the Biolibrarian proposal
Biolibrarians are:
Local
Global
Non-partisan with respect to database or data type.
Non-partisan with respect to software
Pro open source
Pro open access
Pro standards
Pro documentation
Active
Existing problems addressed by the Biolibrarian proposal
Ownership of biological data tranfer
Overwhelmingly large number of biological databases
Absence of stable funding for biological databases curation
The proposal in brief
Time plan: 5 years
Personnel: 8 – 10 biolibrarian curators
Deliverable: A prototype for biolibrarian positions at universities around the world.
We will propose a team of 8 to 10 curators who will search primary biomedical research literature and enter biomolecular interaction and pathway data into machine readable format to facilitate exchange and integration of data with other similar efforts as well as to facilitate human and machine based data mining. These personnel will be trained in the use of the latest pathway, complex, interaction, model organism and protein databases. They will act as a liason between researchers and databases to facilitate retrieval of information AND entry of curated information by local researchers.
Biomolecular interaction data consists of the set of experimentally verified interactions that occur between proteins, DNA’s, RNA’s, small molecules or complexes involving any of these molecular types. These data, along with associated reactions and state changes form the basis of biological pathways. As such, interaction and pathway data define the biological function of their participant molecules. The resulting network of interactions and pathways between molecules forms a map of living systems that may be searched and computed on. The resource is a broadly applicable to all molecular and medical life sciences.
Presently these data are collected by several small groups around the world. It is a labour intensive task that requires skill in reading research articles and knowledge of multiple standard data formats using large controlled vocabularies. Traditionally, these databases have had difficulties in securing long-term, stable funding since they compete with proposals for experimentalist research while they are essentially infrastructure projects. Despite the fact that these databases receive hundreds of citations per year, a survey of the major interaction databases indicates that they employ only a handful of full-time curators. This number is insufficient to keep up with the rate of research publications let alone the backlog of uncurated research articles. This infrastructure call represents a unique opportunity for Norway to establish a prototype position in this area that could be replicated across universities. The cost is fractional compared to the funds expended by universities on biomedical journal subscriptions. The payoff is a powerful dataset that may be data mined by humans and machine algorithms.
We will solicit letters of support from international interaction databases supporting our efforts in this area. We will also solicit national and international research groups to propose biomedical areas requiring curation. Finally, we will survey universities worlwide to assess their support of such a service. Newly curated data will be used to give context to high and low throughput proteomics and sequencing projects as well as provide tools for genome wide analysis studies related to human disease, cancer and personalized medicine. Graphical algorithms acting on large scale interaction and pathway maps have broad utility that includes (but is not limited to) identification of biological roles of proteins, identification of disease genes and selection of drug targets. The efficacy of these algorithms is dependent on the quality of the underlying data. Presently, high-quality, human curated data is dwarfed by less reliable data from high-throughput interactomics studies. The interpretation of these high-throughput studies themselves are benefited by the presence of human-curated data.
The proposed project will have high visibility and high impact. Data will be made freely available in internationally recognized formats (such as the HUPO PSI-MI standard) under a Creative Commons License. Data will be available via bulk-download, web-interface and at least one internationally recognized graphical viewer (http://cytoscape.org). Data will be integrated and exchanged with other similar database efforts to facilitate search and analysis. Integration will be accomplished using a system recently developed in the principal investigator’s research group (http://irefindex.uio.no). This same system will be used to monitor and ensure accepted curation practices. We will contribute to the maintenance and expansion of data exchange formats and controlled vocabularies. We will adhere to and develop curation practices set out by the International Molecular Exchange Consortium (http://imex.sourceforge.net/). Existing curation and database systems for handling data are already available from other IMEx groups and these will be installed, used and built upon.
This initiative is a proposed infrastructure project at the University of Oslo in Norway where this position type would be prototyped. The initiative is led by Ian Donaldson at the Biotechnology Centre of Oslo. The above survey is an attempt to assess support for the proposal at the University of Oslo and at Universities around the world where this project could be replicated.
Ian Donaldson was a lead bioinformatics developer and research scientist for the Biomolecular Interaction Network Database (BIND) between 2002 and 2005. This effort employed close to 30 curators. He was involved in many aspects of this project (including curation and data standard development) since the project’s inception in 1999.
How to add your university to this survey
We have sent query emails to lists of biology researchers at a number of universities. If you would like to do the same for your university, you can use the following message. Simply copy and paste. Results of the survey will be posted on this site.
Subject line: would you like to meet with a biolibrarian?
We are proposing the creation of a new infrastructure position at university libraries around the world. The position is called a “Biolibrarian”.
A Biolibrarian is trained in the use of biological databases that include biological pathway, complex and interaction databases.
It is envisioned that molecular biologists could meet with a Biolibrarian in the same way that they meet with and use the services of a librarian. The Biolibrarian could help molecular biologist researchers to locate pathways, complexes and interactions that their molecules of interest are involved in. The Biolibrarian would help the biologist to access, use and interpret data from curated molecular databases (including pathway, complex, interaction, model organism and protein databases).
Finally, Biolibrarians could help researchers enter verified information from full-text articles into curated databases where it would be available to researchers around the world that were querying for information on these same molecules.
Would you support such a service at your local university library? Do you have comments on this proposal? Please take our six question survey to enter your opinion.
http://www.surveymonkey.com/s.aspx?sm=LubRkhzbX9a7e8aPGKGevQ_3d_3d
You can read more about the proposal here: http://irefindex.uio.no/wiki/The_Biolibrarian_Proposal