Difference between revisions of "iRefScape advanced search"
(New page: ==Path finding: How to find sequences of interactions connecting two proteins== iRefScape could be used to find interaction events connecting two proteins or a sequence of events involving...) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 11: | Line 11: | ||
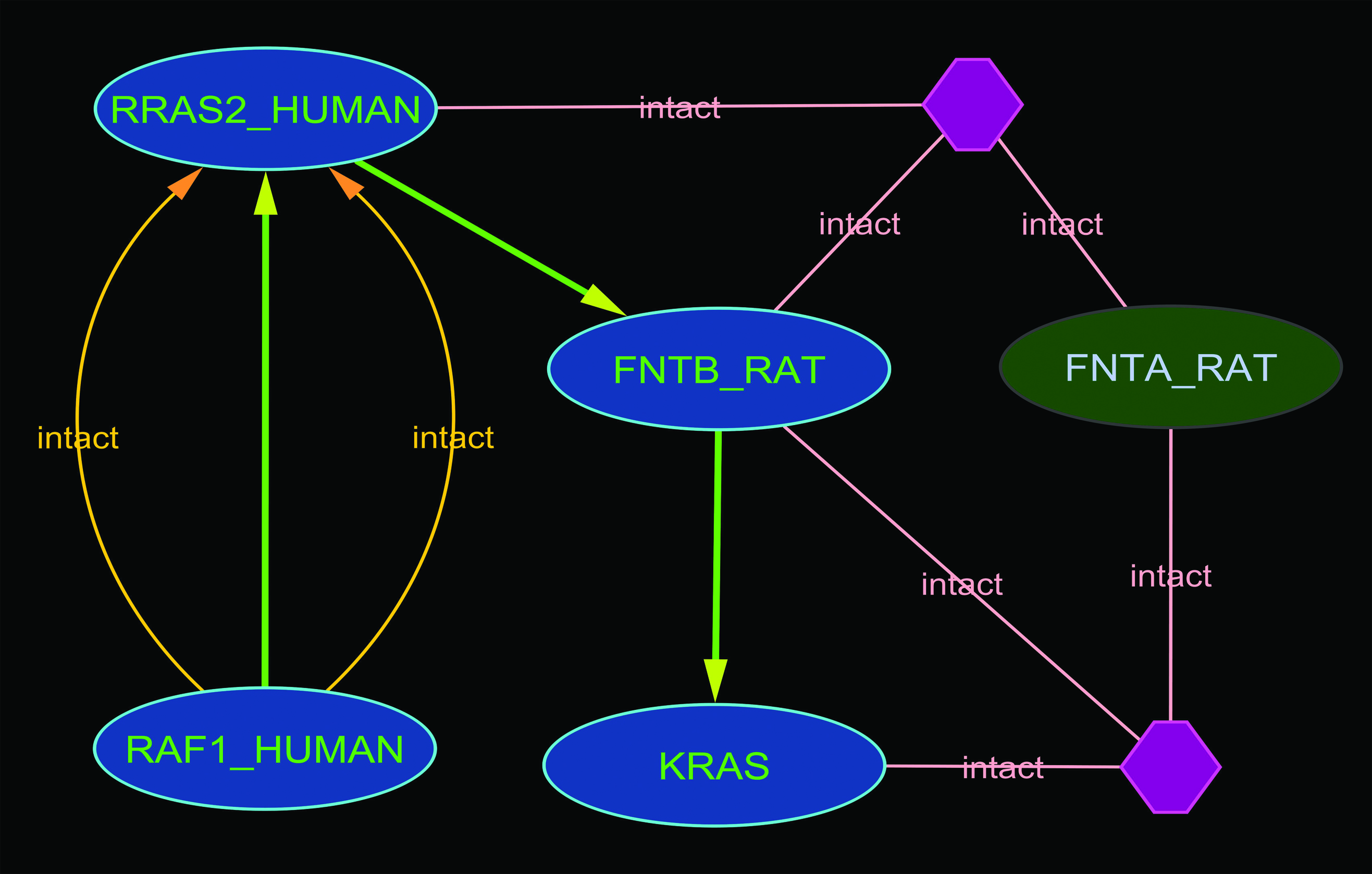
The results of the path finding was sorted in the ascending order of path length and the maximum path length was restricted by the “maximum_distance_allowed” variable with a default value of 6 (the value of this variable could be modified by changing the value of “Maximum distance” from “Path Parameters tab” in advanced options). The paths found in this way were “reasonable paths”, this concept is different from finding the shortest path or finding all the paths. A “reasonable paths” path from A to B is a path extending from A to B where none of the intermediate points can be reached from A with fewer steps by a path that extends from A via B (i.e when evaluating a path from A to B , nodes beyond B are not con-sidered). Due to the way the algorithm works when the opposite search (NP_002871<==>NP_004976) was performed and it took 120 seconds and returned 3 paths. The reason behind this was that NP_002871 is directly connected to many hubs and therefore, initial seed-list size was larger than when stating with NP_004976. The algorithm first try the direction requested by the user and after those considerers the opposite direction when creating the sub-network to look for path. The time consuming step in the NP_002871<==>NP_004976 was the initial evaluation step. When constructing the query for path finding we used the attribute “i.overall_degree_TOP” and it provided the overall connectivity of the proteins within iRefIndex. The “i.overall_degree_TOP” value of RAF1 was 443 and for interested KRAS isoform was 2, therefore, although not shown in the graph RAF is connected to more than 400 other proteins than RAS1. After the path finding was completed the “Path selection” panel can be used to selectively load the paths. In order to make the selection easier, the paths found can be represented by many attribute type and after conversion process the mouse over popup menu displays the path in selected attribute type. | The results of the path finding was sorted in the ascending order of path length and the maximum path length was restricted by the “maximum_distance_allowed” variable with a default value of 6 (the value of this variable could be modified by changing the value of “Maximum distance” from “Path Parameters tab” in advanced options). The paths found in this way were “reasonable paths”, this concept is different from finding the shortest path or finding all the paths. A “reasonable paths” path from A to B is a path extending from A to B where none of the intermediate points can be reached from A with fewer steps by a path that extends from A via B (i.e when evaluating a path from A to B , nodes beyond B are not con-sidered). Due to the way the algorithm works when the opposite search (NP_002871<==>NP_004976) was performed and it took 120 seconds and returned 3 paths. The reason behind this was that NP_002871 is directly connected to many hubs and therefore, initial seed-list size was larger than when stating with NP_004976. The algorithm first try the direction requested by the user and after those considerers the opposite direction when creating the sub-network to look for path. The time consuming step in the NP_002871<==>NP_004976 was the initial evaluation step. When constructing the query for path finding we used the attribute “i.overall_degree_TOP” and it provided the overall connectivity of the proteins within iRefIndex. The “i.overall_degree_TOP” value of RAF1 was 443 and for interested KRAS isoform was 2, therefore, although not shown in the graph RAF is connected to more than 400 other proteins than RAS1. After the path finding was completed the “Path selection” panel can be used to selectively load the paths. In order to make the selection easier, the paths found can be represented by many attribute type and after conversion process the mouse over popup menu displays the path in selected attribute type. | ||
| + | In the image below the green arrow show the path. | ||
| + | |||
| + | <div > | ||
| + | <imagemap> | ||
| + | Image:Path NP 004976 NP 002871 copy.jpg|480x480px | ||
| + | default [http://www.biotek.uio.no] | ||
| + | </imagemap> | ||
| + | </div> | ||
Latest revision as of 16:14, 27 April 2010
Path finding: How to find sequences of interactions connecting two proteins
iRefScape could be used to find interaction events connecting two proteins or a sequence of events involving several proteins.
Path finding connecting two terminal nodes
This process intakes two terminal nodes as input and returns all reasonable paths connecting these two. The results returned here are path way independent. i.e. the sequences of interactions connecting the nodes are not constructed using currently published pathways. However, the paths retuned may contain pathway centric information.
The query format is : NP_004976 <==> NP_002871 . additional type and taxon parameters were also supplied as required. (Type : RefSeq_AC, Taxon:9606). This query located all reasonable paths between NP_004976 and NP_002871 and the returned path also contains the shortest path between them.
The results of the path finding was sorted in the ascending order of path length and the maximum path length was restricted by the “maximum_distance_allowed” variable with a default value of 6 (the value of this variable could be modified by changing the value of “Maximum distance” from “Path Parameters tab” in advanced options). The paths found in this way were “reasonable paths”, this concept is different from finding the shortest path or finding all the paths. A “reasonable paths” path from A to B is a path extending from A to B where none of the intermediate points can be reached from A with fewer steps by a path that extends from A via B (i.e when evaluating a path from A to B , nodes beyond B are not con-sidered). Due to the way the algorithm works when the opposite search (NP_002871<==>NP_004976) was performed and it took 120 seconds and returned 3 paths. The reason behind this was that NP_002871 is directly connected to many hubs and therefore, initial seed-list size was larger than when stating with NP_004976. The algorithm first try the direction requested by the user and after those considerers the opposite direction when creating the sub-network to look for path. The time consuming step in the NP_002871<==>NP_004976 was the initial evaluation step. When constructing the query for path finding we used the attribute “i.overall_degree_TOP” and it provided the overall connectivity of the proteins within iRefIndex. The “i.overall_degree_TOP” value of RAF1 was 443 and for interested KRAS isoform was 2, therefore, although not shown in the graph RAF is connected to more than 400 other proteins than RAS1. After the path finding was completed the “Path selection” panel can be used to selectively load the paths. In order to make the selection easier, the paths found can be represented by many attribute type and after conversion process the mouse over popup menu displays the path in selected attribute type. In the image below the green arrow show the path.