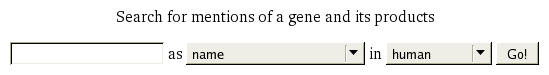Difference between revisions of "Using the Bioscape Web Application"
PaulBoddie (talk | contribs) (Moved images.) |
PaulBoddie (talk | contribs) (Added category.) |
||
| Line 152: | Line 152: | ||
* When searching by <tt>Database plus identifier</tt>, the [[#Navigating_Document_Versions|versions of the source document]] will be presented, should the identifier be considered valid. | * When searching by <tt>Database plus identifier</tt>, the [[#Navigating_Document_Versions|versions of the source document]] will be presented, should the identifier be considered valid. | ||
* When searching by <tt>Bioscape identifier</tt>, the exact document will be displayed, should the identifier be valid. | * When searching by <tt>Bioscape identifier</tt>, the exact document will be displayed, should the identifier be valid. | ||
| + | |||
| + | [[Category:Bioscape]] | ||
Revision as of 17:07, 24 July 2009
The Bioscape Web application seeks to provide a convenient interface to textual documents which have been searched for bioentity (gene and/or protein) information, as well as providing an overview of each bioentity known to the system.
Contents
Searching
The start page for Bioscape provides a minimal search dialogue where names or identifiers of bioentities can be specified, together with the kind of bioentity being searched for, and along with any organism-related restrictions. Upon searching (by pressing the Go! button), if only a single gene satisfies the search criteria, a summary page will be shown for that gene; otherwise, a disambiguation page will be shown first, as described in "Search Disambiguation".
Searching for Genes by Name
An example:
- Enter LHbeta into the text box.
- Select name from the input type menu.
- Select either human or all organisms from the organism menu, according to preference.
- Press the Go! button.
Bioscape will not only attempt to match the given name against the names recorded for genes in typical gene databases, but it will also attempt to suggest genes that are thought to employ the given name in the searched literature. Consequently, for the above example, genes known by the names LH2 and LHB will be suggested. (This depends on a technique known as orthographic variation.)
Searching for Genes by Identifier
An example:
- Enter 1234 into the text box.
- Select Entrez Gene identifier from the input type menu.
- Select either human or all organisms from the organism menu, according to preference.
- Press the Go! button.
Bioscape will attempt to show a summary page for the chosen gene. If the chosen organism is not compatible with the gene whose identifier has been given, or if no such identifier is known, an error will be shown.
Searching for Genes via Protein Accessions
An example:
- Enter NP_001099006.1 into the text box.
- Select RefSeq protein accession from the input type menu.
- Select either human or all organisms from the organism menu, according to preference.
- Press the Go! button.
Bioscape will attempt to show the summary page for the gene associated with the specified product.
Search Disambiguation
When Bioscape cannot be sure which gene whose details the user wishes to investigate, a disambiguation page will appear and the user will be requested to choose one of a number of suggestions.
An example:
- First, search for LHbeta as a name in human (as described in "Searching for Genes by Name").
- A number of suggestions are proposed for genes in which the user might be interested.
- Press the Go! button next to the gene which is most appropriate, such as LH2 ("luteinizing hormone beta polypeptide", Entrez Gene #3972).
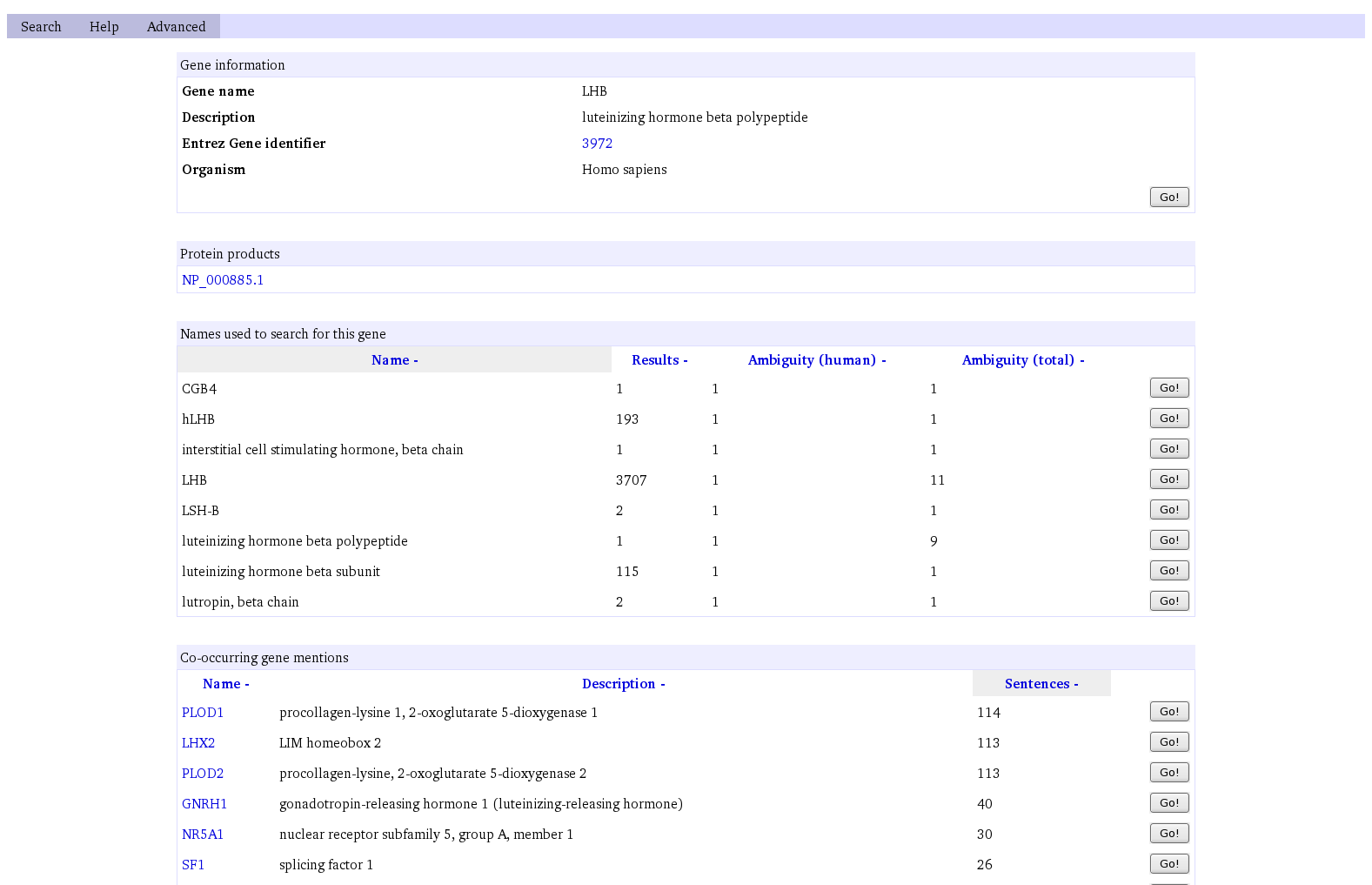
Gene Summaries
Each gene summary contains the following sections:
- Basic information about the gene.
- A list of protein products associated with the gene.
- A list of names by which the gene is known.
- A list of co-occurrences showing, for the featured gene, those genes which appear together with the gene in sentences in the literature.
From most sections, a Go! button is provided which leads to textual results showing "mention sentences" involving the gene:
- From the basic information, textual results show any mention of the gene under any name.
- From the list of names, textual results show only mentions of the gene using the corresponding specific name.
- From the list of co-occurrences, textual results show any co-occurring mention of the two genes concerned (the featured gene and that given in the list) under any of their names.
Mention Sentences
The following kinds of mention sentence pages are provided in Bioscape:
- Mentions of a gene under any name.
- Mentions of a gene using a specific name.
- Mentions of a gene appearing together with another specified gene, for any known name employed by either gene.
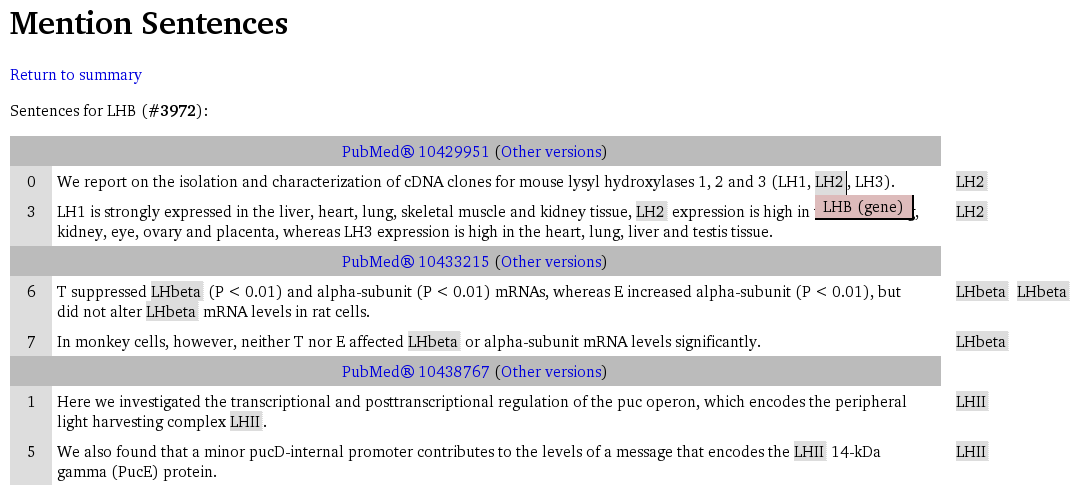
In all such pages, a list of sentences grouped by document are shown, and the following features are provided:
- For each document, a header shows the source of the document, providing a link to the complete document summary and a link to a list of document versions (described below).
- Each document provides a number of sentences featuring the gene as described above. Each mention is highlighted showing elementary details of the mention; for more details the complete document summary should be visited.
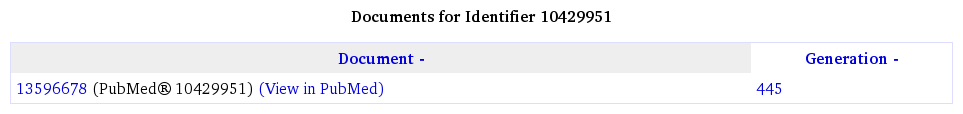
Each source document imported into Bioscape may be indexed many times, either to implement different tokenisation schemes or to provide different versions of the document. A page showing the versions of a document may be viewed by following links labelled with text resembling "other versions".
Each entry in a list of document versions shows the following information:
- The Bioscape document identifier.
- The original source document identifier.
- A generation number indicating the collection (or batch) which provided this version of the document.
It may be useful to refer to a specific document version, referenced by a Bioscape document identifier and optionally a generation number, in order to examine specific search results and annotations. In general, however, it is typically more useful to refer to source document identifiers together with the database which provided the document to Bioscape.
Document Summaries
The following kinds of document summaries are provided by Bioscape:
- Original document (index) summaries show the unannotated text for each document, split into sentences and with any relevant metadata.
- Search result document (unscored) summaries show the text for each document with textual search results highlighted.
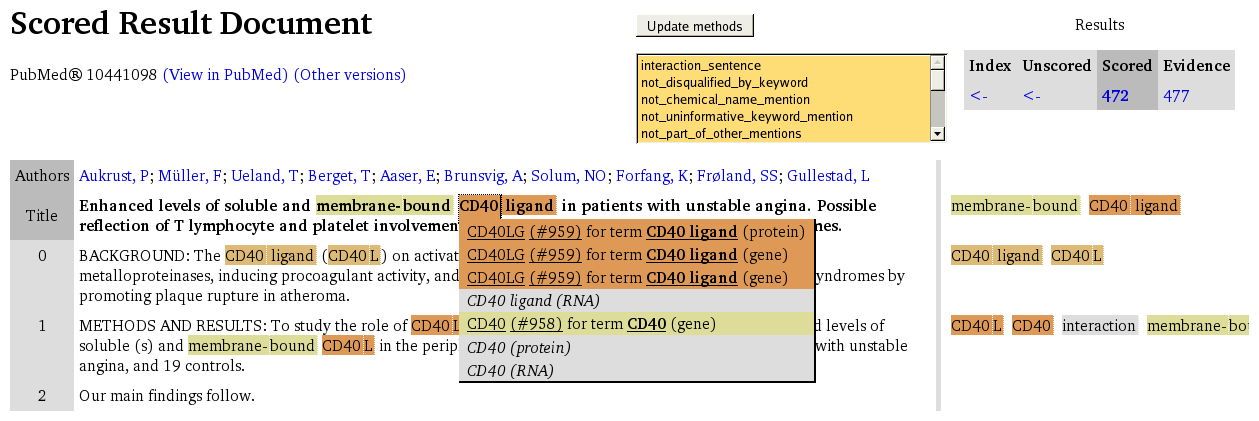
- Scored result document summaries show the text with specific (or concrete) search results highlighted.
- Interaction evidence document summaries show the text with results highlighted on sentences which are thought to provide interaction evidence.
The highlighting of results varies according to the kind of summary, but each highlighted region produces a pop-up element showing specific information about the highlighted region:
- Search result document (unscored) summaries provide details of the names or terms which were found.
- Scored result document summaries provide details of the bioentities found.
- Interaction evidence document summaries provide suggestions of possible interactions using pairwise combinations of result bioentities.
Clicking on highlighted regions should fix pop-up elements in an open state, and if script support is enabled in the browser, it should be possible to keep more than one pop-up element in such a "fixed open" state at any given time; otherwise, only one pop-up element may be kept open in this way, although other elements may be opened by hovering over them with the pointer.
Within each pop-up element, a range of links to other resources are typically provided:
- Links to Entrez Gene pages about each gene.
- Links to scoring information about a particular gene mention.
- Links to scoring information about a particular name mention.
Scoring and Methods
In scored result and interaction evidence summaries, the effect of scoring can be observed on the results. It is possible to select a combination of scoring methods from those available for the results and to see the resulting assessment of those results; this is done by using the method selection list and by pressing the Update methods button.
Each region in a scored summary is coloured according to the best scoring mention at that precise location, where the range of colours from lowest to highest score is given in the legend which appears on each summary page of this nature. Within the pop-up element for each region, there should be a list of mentions together with details of the suggested bioentity, coloured according to the assessment of the candidate's suitability made by the combination of methods selected from the list described above.
It should be noted that some methods will always contribute positive assessments because they have been used to filter results. Thus, selecting them will not yield a mixture of positive and negative assessments amongst the results - in effect, they should behave in a neutral fashion. The contributions made by each method can be studied by following the links to scoring information provided in each pop-up element.
Advanced Search
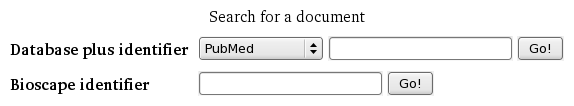
The Advanced navigation link may be followed to reveal the advanced search dialogue provided by Bioscape. This screen can be used to find documents according to their source database identifier or using a specific Bioscape identifier (as described in the "Navigating Document Versions" section).
- When searching by Database plus identifier, the versions of the source document will be presented, should the identifier be considered valid.
- When searching by Bioscape identifier, the exact document will be displayed, should the identifier be valid.